Perfusion quantification in Tumours using Multi-PLD pCASL¶
The purpose of this exercise is to look at some multi-PLD pcASL in a clinical example of glioblastoma multiforme [1] [2] to assess how perfusion changes within the tumour.
Contents
Basic Orientation¶
Before we do any data processing, this is a quick orientation guide to Quantiphyse if you’ve not used it before. You can skip this section if you already know how the program works.
Start the program by typing quantiphyse at a command prompt, or clicking on the Quantiphyse
icon  in the menu or dock.
in the menu or dock.
Loading the data¶
If you are taking part in an organized practical workshop, the data required may be available in your home
directory, in the course_data/ASL_IMAGO folder. If not, an encrypted zipfile containing the data can be
downloaded below - you will be given the password by the course organizers:
Note
To extract the 7zip archive on Linux, download and then use the command 7z x IMAGOASL_Michigan.7z
Start by loading the ASL data into Quantiphyse - use File->Load Data or drag and drop to load
the file mpld_asltc.nii.gz. In the Load Data dialog select Data.
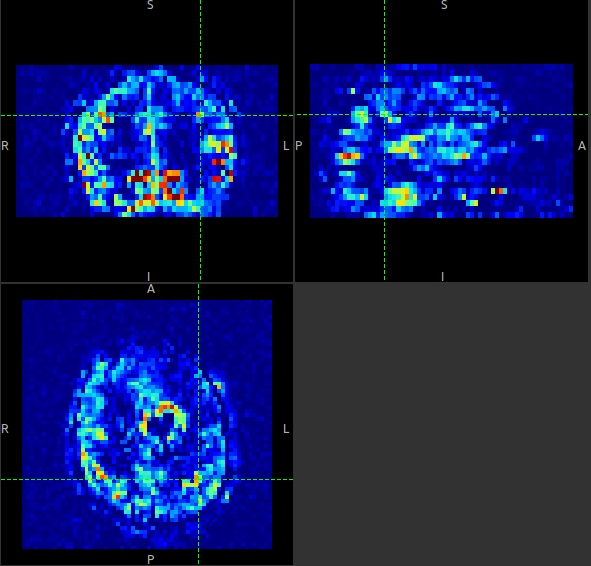
The data should look as follows:
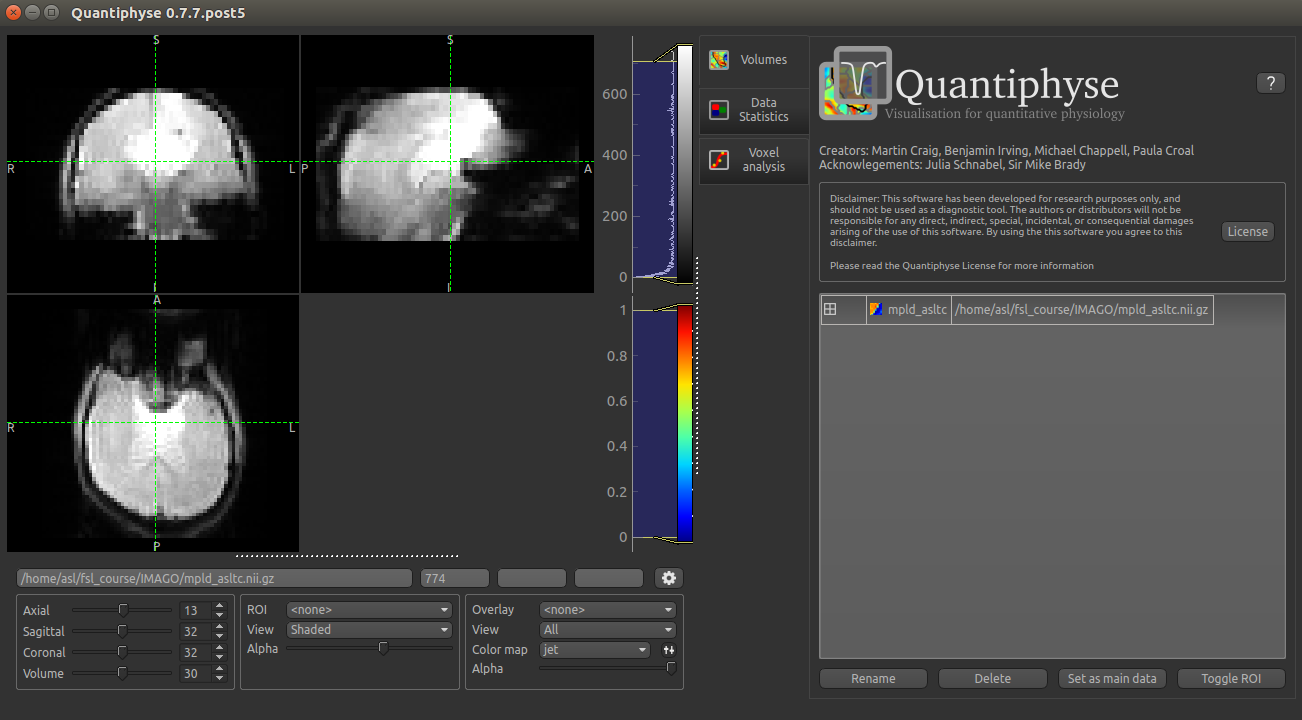
Image view¶
The left part of the window contains three orthogonal views of your data.
- Left mouse click to select a point of focus using the crosshairs
- Left mouse click and drag to pan the view
- Right mouse click and drag to zoom
- Mouse wheel to move through the slices
- Double click to ‘maximise’ a view, or to return to the triple view from the maximised view.
Widgets¶
The right hand side of the window contains ‘widgets’ - tools for analysing and processing data. Three are visible at startup:
Volumesprovides an overview of the data sets you have loadedData statisticsdisplays summary statistics for data setVoxel analysisdisplays timeseries and overlay data at the point of focus
Select a widget by clicking on its tab, just to the right of the image viewer.
More widgets can be found in the Widgets menu at the top of the window. The tutorial
will tell you when you need to open a new widget.
For a slightly more detailed introduction, see the Getting Started section of the User Guide.
Perfusion quantification¶
In this section we will quantify perfusion for the dataset just loaded.
This dataset used pCASL labelling, with a duration of 1.8 seconds, and 5 post-labeling delays of 0.4, 0.8, 1.2, 1.6 and 2.0 seconds. The label-control ASL series contains 60 volumes, with each PLD repeated 6 times, thus there are 12 volumes (label and control paired) each PLD. The data is in the order that it was acquired, which will be important for setting up the analysis.
A perfusion weighted image¶
Open the Widgets->ASL->ASL Data Processing widget. We do not need to set all the details of the
data set yet, however note that the data format is (correctly) set as Label-control pairs.
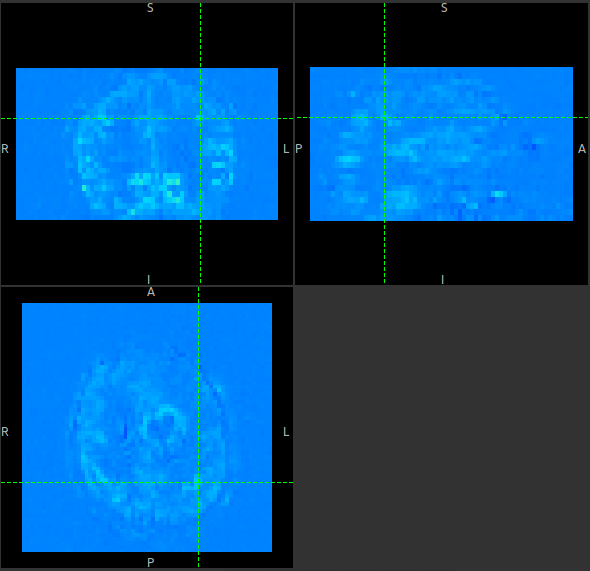
Click on the Generate PWI button. This performs label-control subtraction and averages the
result over all repeats. The result is displayed as a colour overlay, which should look like a
perfusion image:
We can improve the display a little by adjusting the colour map. Find the overlay view options below the main image view:
Next to the Color Map option (which you can change if you like!) there is a levels button  which lets you change the min and max values of the colour map. Set the range from
which lets you change the min and max values of the colour map. Set the range from 0 to 10
and select Values outside range to Clamped.
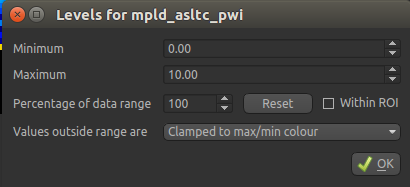
Then click Ok. The perfusion weighted image should now be clearer:
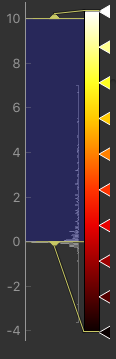
Colour map widget
You could also have modified the colour map limits by dragging the colourmap range widget directly - this is located to the right of the image view. You can drag the upper and lower limits with the left button, while dragging with the right button changes the displayed scale. You can also customize the colour map by clicking on the colour bar with the right button.
Warning
Dragging the colourmap is a little fiddly due to a GUI bug. Before trying to adjust the levels, drag down with the right mouse button briefly on the colour bar. This unlocks the automatic Y-axis and will make it easier to drag on the handles to adjust the colour map.
Model based analysis - Data set up¶
Looking at the ASL data processing widget we used to generate the PWI, you can see that this
is a multi-page widget in which each tab describes a different aspect of the analysis pipeline.
We start by inputing the information on the first page which describes the ASL data. The defaults are shown below but we will need to change some of them to correctly describe our ASL acquisition.
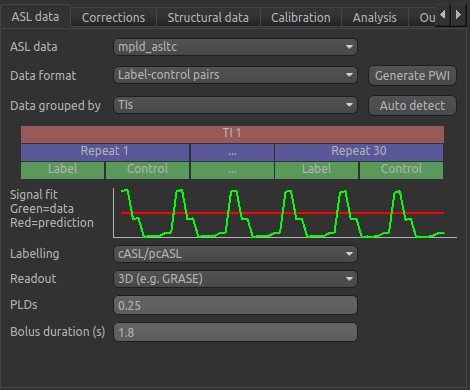
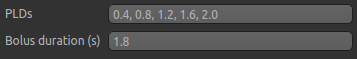
Firstly, we need to enter the 5 PLDs in the PLDs entry box – these can be separate by spaces or commas. We also make sure the label duration is set to 1.8s:
The data was acquired in label-control pairs (the default setting), and grouped by repeats. We need
to change the Data Grouped By option to Repeats to reflect this. Below this selection there
is a graphical illustration of the structure of the data set:
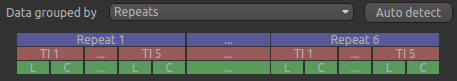
The data set volumes go from left to right. Starting with the top line (blue) we see that the data set consists of 6 repeats, and within each repeat there are 5 TIs (red), each with a label and control image (green).
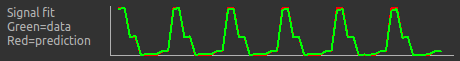
Below the grouping diagram, there is a visual preview of how well the actual data signal matches what would be expected from this grouping. The actual data signal is shown in green, the expected signal from the grouping is in red, and here they match nicely, showing that we have chosen the correct grouping option.
If we change the Data Grouped By option to TIs (incorrect) we see that the actual and expected signal do not match up:
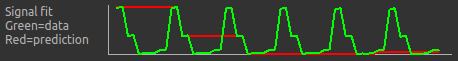
We can get back to the correct selection by clicking Auto Detect which chooses the grouping which gives
the best match to the signal.
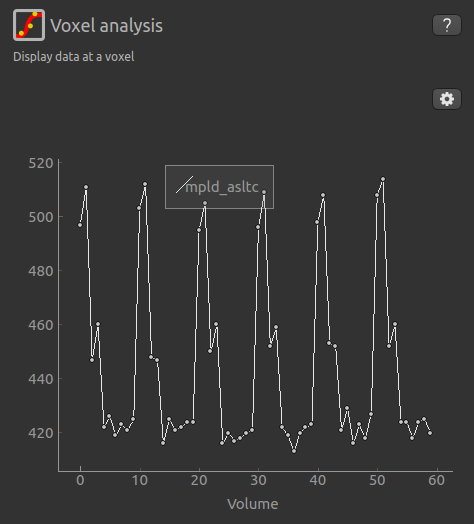
Another way to determine the data ordering is to select the Voxel Analysis widget and click on a GM
voxel, which should clearly show 6 groups of repeats. Each of the 6 peaks represents a single repeat
across all 5 PLDs, the zig-zag pattern of the label-control images are visible for each PLD.
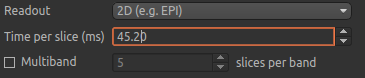
Returning to the ASL data processing page, we need to finalise our acquisition details. The labelling method is correctly set to cASL/pCASL, however we have a 2D readout with 45.2 ms between slices, so we need to change the Readout option to reflect this. When we select a 2D readout, the option to enter the slice time appears automatically.
Model based analysis - Analysis set up¶
In this section we invert the kinetics of the ASL label delivery to fit a perfusion image, and use the calibration image to get perfusion values in the units of ml/100g/min.
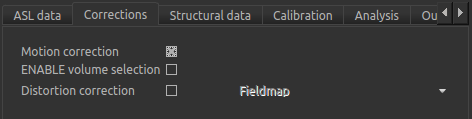
Firstly, on the Corrections tab, we will ensure that Motion Correction is checked (this should
be enabled by default):
Due to potential challenges with MNI registration in the presence of tumour, we will work in the
subject’s native space, thus skip the Structural data tab, and instead move on to Calibration. To
use calibration we first need to load the calibration image data file from the same folder containing
the ASL data - again we can use drag/drop or the File->Load Data menu option to load the
following files:
aslcalib.nii.gz- Calibration (M0) imagecsfmask.nii.gz– CSF mask in subject’s native space
Note
For the csfmask data ensure that this is loaded as an ROI in the data type selection box. If you
forget to do this, you can modify it from the Volumes widget - click on the data set in the list
and click the Toggle ROI button.
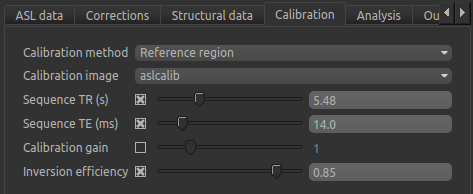
On the Calibration tab we will set the calibration method as Reference region, and will need to select
the calibration image we have just loaded: aslcalib. The TR for this image was 5.48s, so click on the
Sequence TR checkbox and set the value to 5.48. Similarly, click on the Sequence TE checkbox and set the
value to 14.0. Finally, change the Inversion efficiency to 0.85 as we are using pCASL (the
GUI is set to the PASL default of 0.98).
In the Reference region calibration box we will select the CSF
option, and set the Custom reference ROI to the csfmask ROI which we have just loaded.
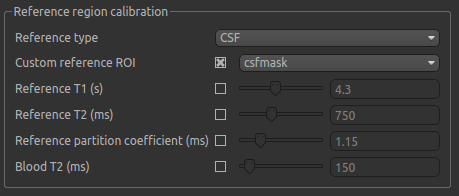
In the interest of time, this CSF mask has been made manually ahead of time, and provides a conservative mask within the ventricles.
On the Analysis tab the defaults do not need altering in this instance, except to turn the macrovascular component
off.
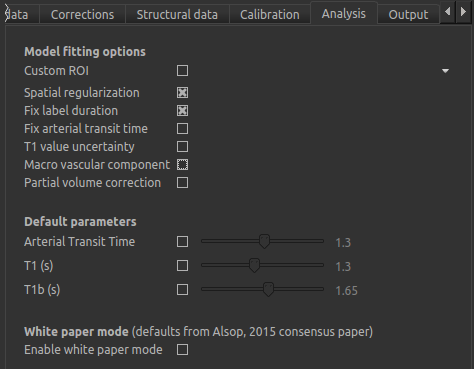
We will not change the defaults on the Output tab yet, but will select Save HTML report. Click
Choose to set the output folder.
We are now set up to run the analysis - but before you do, check the green box at the bottom of
the widget which reports where it thinks FSL is to be found. If the information does not seem
to be correct, click the Change button and select the correct location of your FSL
installation (if you are in an organized practical this should be correct).

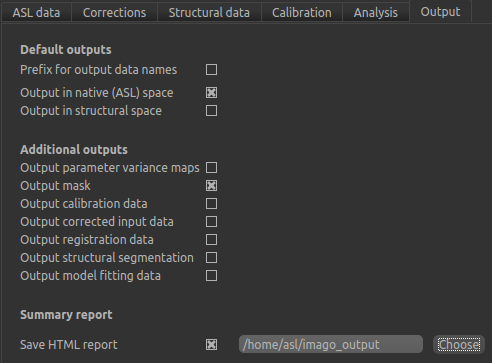
As an additional step, you may want to save your output data. You can of course save the output data from your analysis
after it has run using File->Save Current Data, however it’s often useful to have all the output saved automatically for
you. By selecting Save copy of output data (underneath the Run button) and choosing an output folder, this will be done.
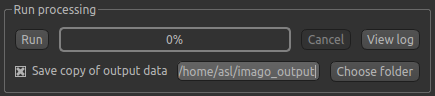
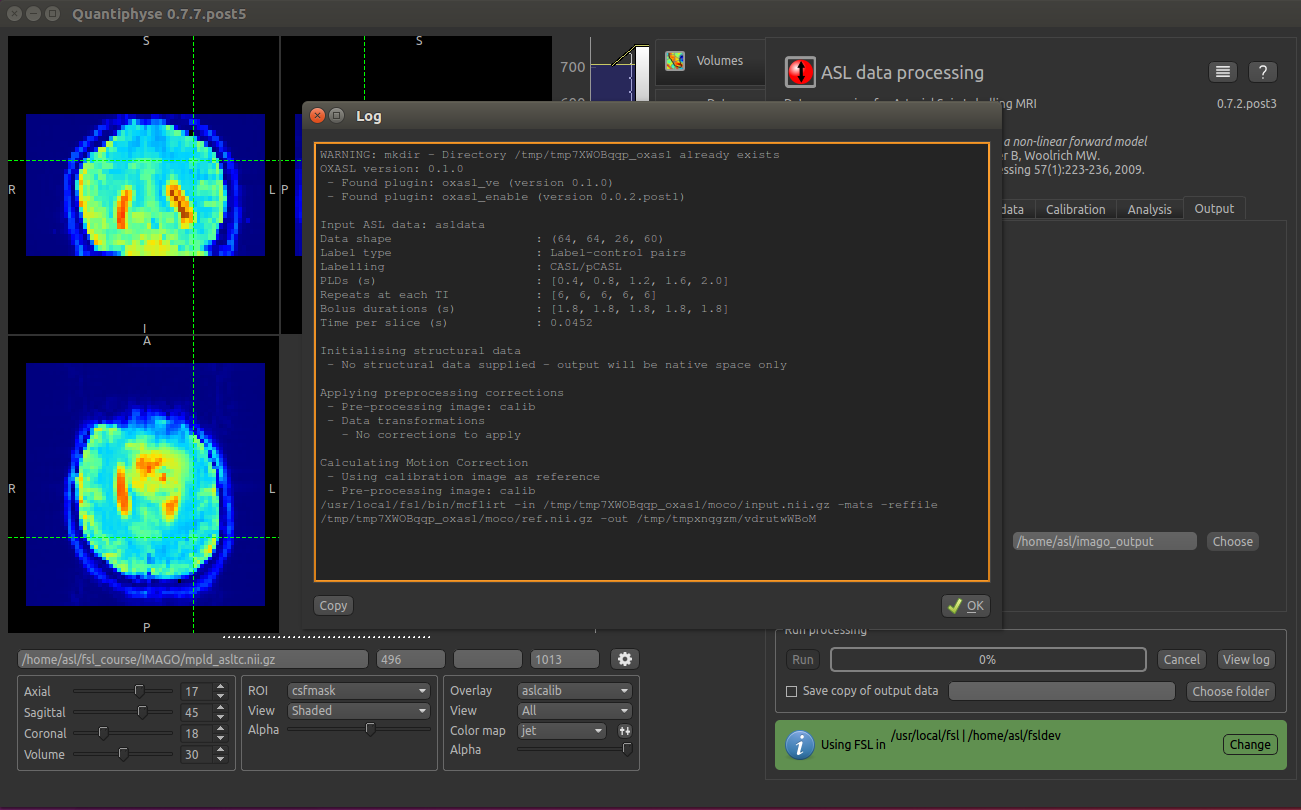
Finally click Run at the bottom to run the analysis. You can click the View Log button
to view the progress of the analysis which should only take a few minutes.
Once the analysis had completed (~5 mins), some new data items will be available. You can display them either
by selecting them from the Overlay menu below the image display, or by clicking on the
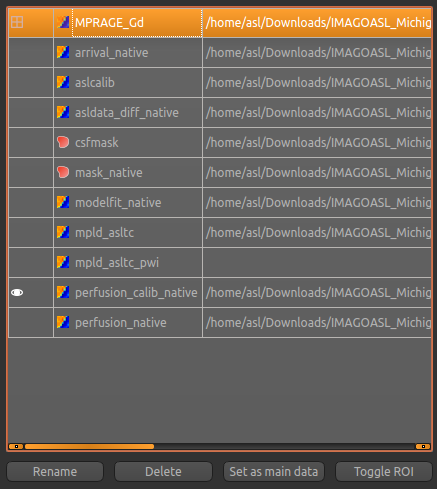
Volumes widget and selecting them from the list. The new data items are:
perfusion_native- Raw (uncalibrated) perfusion mapperfusion_calib_native- Calibrated perfusion data in ml/100g/minarrival_native- time it takes for blood to transit between the labeling and imaging regions.mask_native- An ROI (which appears in the ROI selector under the image view) which represents the region in which the analysis was performed.
We can view these outputs within the brain mask only, by selecting mask_native from the ROI dropdown.
The images may be clearer if we modify the view style for the ROI from Shaded to Contour (in the
ROI options box underneath the image view). This replaces the translucent red mask with an outline:
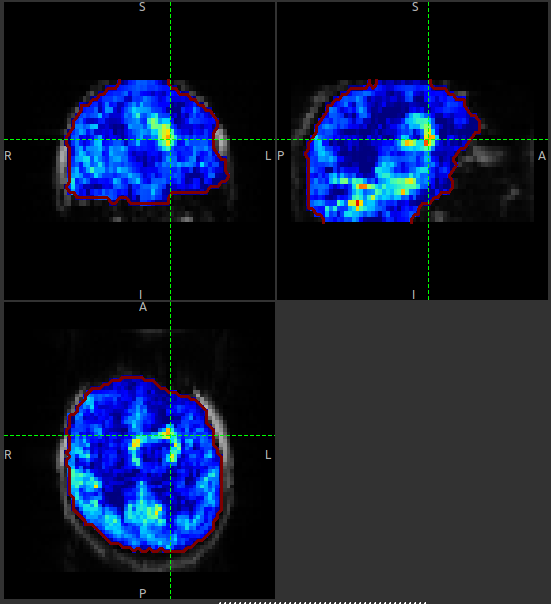
The perfusion_calib_native image should look similar to the perfusion weighted image we created
initially, however the data range reflects the fact that it is in physical units. To get a clear
visualustion set the colour map range to 0 – 60, and clamping to min/max using the Levels button  .
You can also select
.
You can also select Only in ROI as the View option just above this so we only see the perfusion map within the
selected ROI.
The result should look something similar to below. Notice that you can see a ring of perfusion enhancement near the midline, this is consistent with tumour location, and gadolinium enhancement.
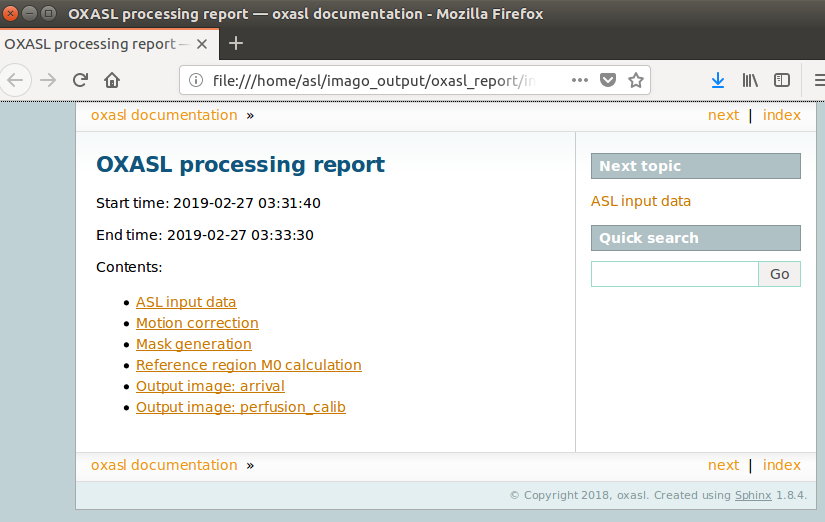
As well as outputting images, Quantiphyse will attempt to open the analysis report in your default web
browser when the pipeline has completed, but if this does not happen you can navigate to the directory
yourself and open the index.html file.
Below is an example of the information included in the report:
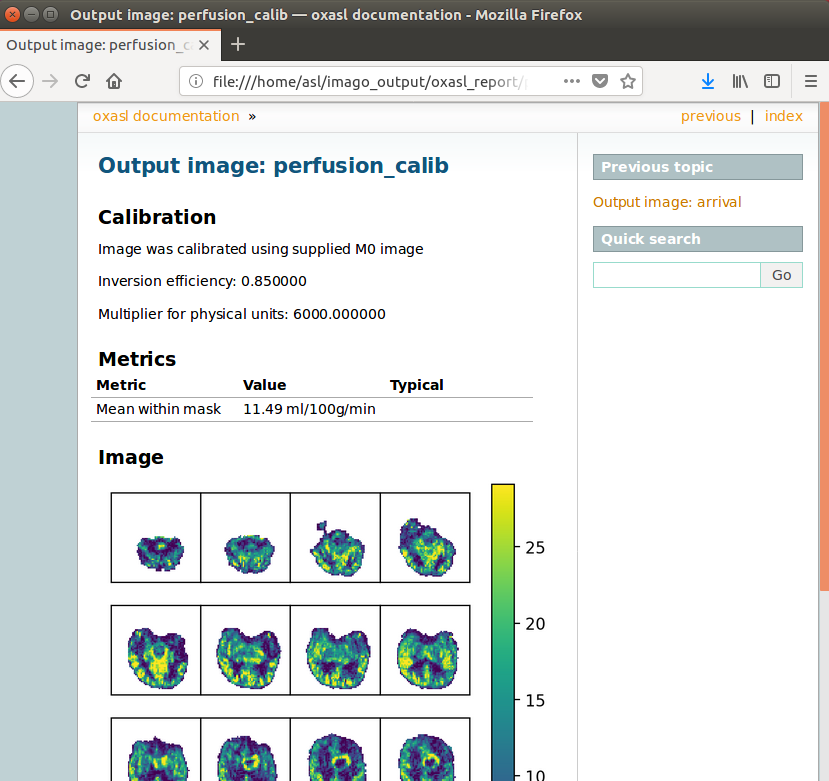
The links are arranged in the order of the processing steps and each link leads to a page giving more detail on this part of the pipeline. For example, if we click on the perfusion image link we get a sample image, which can be to check that the analysis seems to have worked as expected. Here, the mean within mask is not as informative as it might be for a healthy brain, as we are likely averaging in hypoperfused regions.
Comparison to structural changes¶
You may want to see how well the perfusion map corresponds to the tumour visualised on a typical anatomical
image. You can load the patient’s gadolinium-enhanced T1-weighted scan using File->Load Data and
MPRAGE_Gd.nii.gz.
In order to overlay images on top of this structural image, check the Set as main data
box when loading:
Note
If you forget to do this you can also select the Volumes widget, click on the MPRAGE_Gd image and click
the Set as main data button.
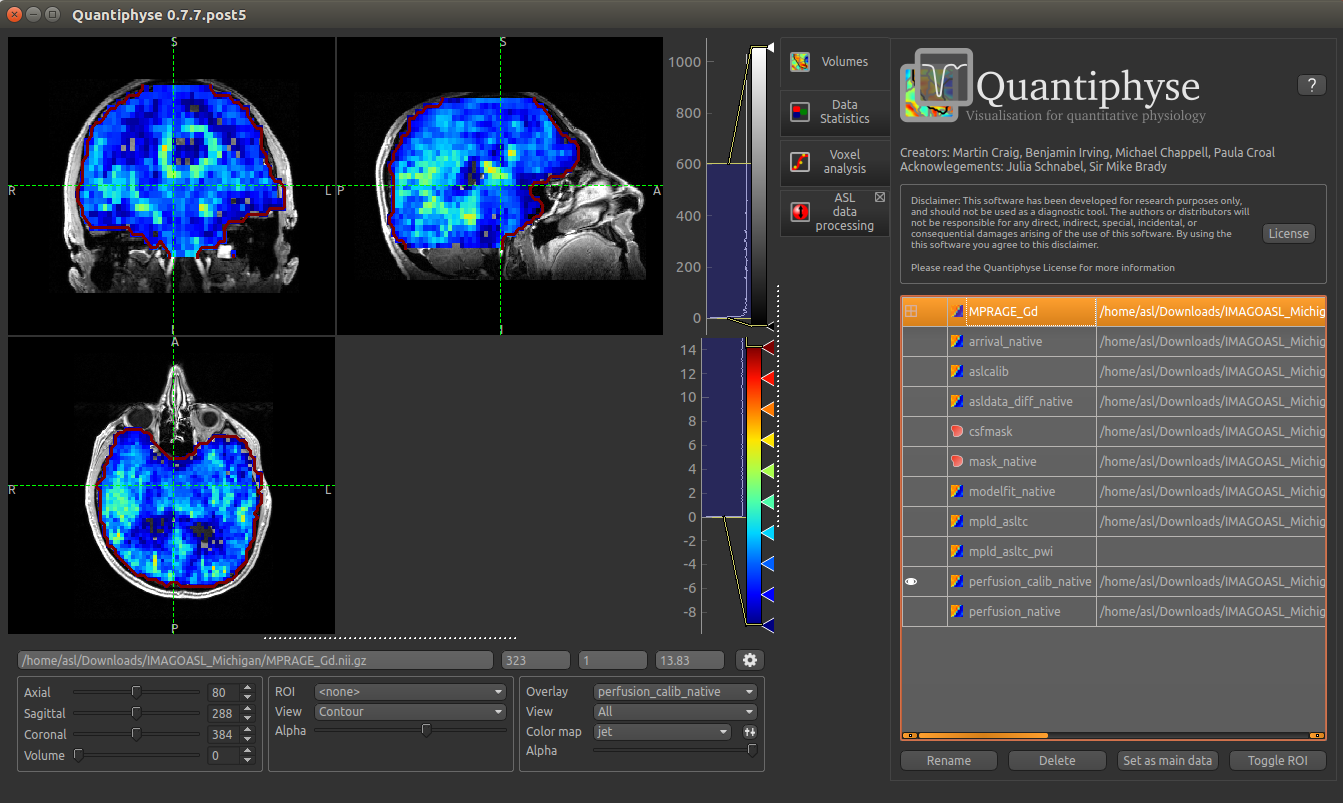
After setting the anatomical image as the main data you, other images selected from the Overlay list
will be overlaid on top, for example the calibrated perfusion map:
The Alpha slider in the overlay box can be used to adjust the transparency of the overlay and compare to
the anatomical image underneath.
You should be able to see that the T1 enhancing rim of the tumour corresponds to a region of increased perfusion. We could go on to load ROI’s of the tumour and contralateral tissue to quantify this, however it is beyond the scope of this tutorial.
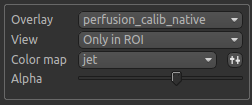
Note
These visualisations work best when Only in ROI is selected as the overlay view option.